3. Registration for location of programs and resources
Page 정보
작성자 TGFam-admin Reply 0건 Read 57,069회 작성일 18-07-18 11:30본문
To run TGFam-Finder, users should prepare RESOURCE.config including full location of genomic resources, and PROGRAM_PATH.config containing absolute location of pre-installed programs and name of output directories. Through the full auto-installation, RESOURCE.config for sample data and PROGRAM_PATH.config are automatically generated. Users who do not want to use our installation script should manually input location of the programs in PROGRAM_PATH.config as well as the resources in RESOURCE.config. If users perform partial-installation except for InterproScan using our script, the location of InterproScan should be inserted in PROGRAM_PATH.config.
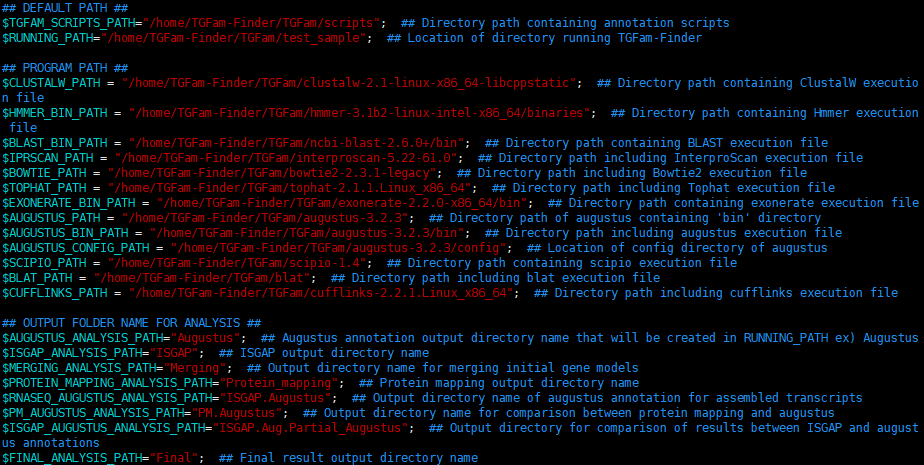
3.1. PROGRAM_PATH.config
## DEFAULT PATH ##
1. $TGFAM_SCRIPTS_PATH=""; ## Directory path containing annotation scripts
2. $RUNNING_PATH=""; ## Location of directory running TGFam-Finder
## PROGRAM PATH ##
1. $CLUSTALW_PATH = ""; ## Directory path containing ClustalW execution file
2. $HMMER_BIN_PATH = ""; ## Directory path containing Hmmer execution file
3. $BLAST_BIN_PATH = ""; ## Directory path containing BLAST execution file
4. $IPRSCAN_PATH = ""; ## Directory path including InterproScan execution file
5. $BOWTIE_PATH = ""; ## Directory path including Bowtie2 execution file
6. $TOPHAT_PATH = ""; ## Directory path including Tophat execution file
7. $EXONERATE_BIN_PATH = ""; ## Directory path containing exonerate execution file
8. $AUGUSTUS_PATH = ""; ## Directory path of augustus containing ‘bin’ directory
9. $AUGUSTUS_BIN_PATH = ""; ## Directory path including augustus execution file
10. $AUGUSTUS_CONFIG_PATH = ""; ## Location of config directory of augustus
11. $SCIPIO_PATH = ""; ## Directory path containing scipio execution file
12. $BLAT_PATH = ""; ## Directory path including blat execution file
13. $CUFFLINKS_PATH = ""; ## Directory path including cufflinks execution file
## OUTPUT FOLDER NAME FOR ANALYSIS ##
1. $AUGUSTUS_ANALYSIS_PATH=""; ## Augustus annotation output directory name that will be created in RUNNING_PATH ex) Augustus
2. $ISGAP_ANALYSIS_PATH=""; ## ISGAP output directory name
3. $MERGING_ANALYSIS_PATH=""; ## Output directory name for merging initial gene models
4. $PROTEIN_MAPPING_ANALYSIS_PATH=""; ## Protein mapping output directory name
5. $RNASEQ_AUGUSTUS_ANALYSIS_PATH=""; ## Output directory name of augustus annotation for assembled transcripts
6. $PM_AUGUSTUS_ANALYSIS_PATH=""; ## Output directory name for comparison between protein mapping and augustus
7. $ISGAP_ AUGUSTUS_ANALYSIS_PATH=""; ## Output directory for comparison of results between ISGAP and augustus annotations
8. $FINAL_ANALYSIS_PATH=""; ## Final result output directory name
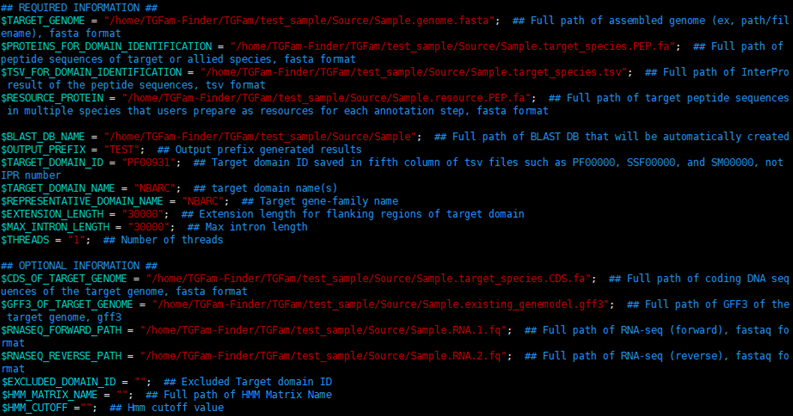
3.2. RESOURCE.config
## REQUIRED INFORMATION ##
1. $TARGET_GENOME = ""; ## Full path of assembled genome (ex, path/filename), fasta format
2. $PROTEINS_FOR_DOMAIN_IDENTIFICATION = ""; ## Full path of peptide sequences of target or allied species, fasta format
3. $TSV_FOR_DOMAIN_IDENTIFICATION = ""; ## Full path of InterPro result of the peptide sequences, tsv format
4. $RESOURCE_PROTEIN = ""; ## Full path of target peptide sequences in multiple species that users prepare as resources for each annotation step, fasta format
5. $BLAST_DB_NAME = ""; ## Full path of BLAST DB that will be automatically created
6. $OUTPUT_PREFIX = ""; ## Output prefix generated results
7. $TARGET_DOMAIN_ID = ""; ## Target domain ID saved in fifth column of tsv files such as PF00000, SSF00000, and SM00000, not IPR number
8. $TARGET_DOMAIN_NAME = ""; ## target domain name(s)
9. $REPRESENTATIVE_DOMAIN_NAME = ""; ## Target gene-family name
10. $EXTENSION_LENGTH = ""; ## Extension length for flanking regions of target domain
11. $MAX_INTRON_LENGTH = ""; ## Max intron length
12. $THREADS = ""; ## Number of threads
## OPTIONAL INFORMATION ##
1. $CDS_OF_TARGET_GENOME = ""; ## Full path of coding DNA sequences of the target genome, fasta format
2. $GFF3_OF_TARGET_GENOME = ""; ## Full path of GFF3 of the target genome, gff3
3. $RNASEQ_FORWARD_PATH = ""; ## Full path of RNA-seq (forward), fastaq format
4. $RNASEQ_REVERSE_PATH = ""; ## Full path of RNA-seq (reverse), fastaq format
5. $EXCLUDED_DOMAIN_ID = ""; ## Excluded Target domain ID
6. $HMM_MATRIX_NAME = ""; ## Full path of HMM Matrix Name, HMM format
7. $HMM_CUTOFF =""; ## Hmm CutOFF value (default is 1e-3)
Note. If you have problems for preparation of config
files, see the section 6.2 and 6.3.
ReplyList
Register된 Reply이 없습니다.

