6. Points to note
Page 정보
작성자 TGFam-admin Reply 0건 Read 12,654회 작성일 18-07-18 11:42본문
6.1 If TGFam-Finder installation is failed
TGFam-Finder is a program based on known bioinformatics programs. Auto installation script is provided to make it easier for the user to install, since the specifications and status of all servers or computers are not the same, sometimes the installation is not perfect and errors may occur.
At this time, check whether the following libraries and modules are properly installed, and then execute the automatic installation again.
Check list
Yum (Ubuntu: apt-get): Use yum to update the following packages
- Packages for rpm or yum-based Linux distributions (RedHat / Fedora / CentOS) are:
zlib-devel, bzip2-devel, xz-devel
- Packages for dpkg-based Linux distributions (Debian /
Ubuntu) are:
zlib1g-dev, libbz2-dev, liblzma-dev
Perl 5.6.1 or higher Version 5.8 or higher is highly recommended. Modules are tested against version 5.8 and above on 5.8 and above.
BioPerl, perl module – yaml, Glib version 3.20 or higher, KentLib, Libpng, Ncurses,
GNU make & C compiler (e.g. gcc or clang), Boost C++ libraries (version 1.47 or higher)
6.2 Configuration
- Users can
input multiple ‘TARGET_DOMAIN_ID’, ‘TARGET_DOMAIN_NAME’ using comma delimiter
but target domain names should be distinct.
ex) $TARGET_DOMAIN_ID = “PF00319,cd00265”
$TARGET_DOMAIN_NAME = “SRF,MEF”
- If users want to obtain final gene model including existing gene model of the target genome, users need to input CDS_OF_TARGET_GENOME and GFF3_OF_TARGET_GENOME.
- IDs in peptides (not RESOURCE_PROTEIN), tsv, coding DNA sequences, and gff3 should be matched. If users use merged peptides and tsv of genomes including target and allied species, IDs of the peptides and tsv should contain IDs of CDS and gff3.
- If users don’t insert RNASeq information, ISGAP and related analyses will not be performed and final gene model is generated combining protein mapping and augustus annotation except for ISGAP and related analyses.
- For annotation of genes having short target domains such as C2H2 zinc finger, we recommend that users register HMM_MATRIX_NANE for hmm matrix for specific domain ID(s). For example, we used PF00096.hmm in Pfam hmm matrix for annotation of C2H2 zinc finger gene families in animal genomes.
6.3 Cautions for preparation of genomic resources
- Gff3 file
should be simplified. Like the capture below, information of coding genes in the
third column should contain gene, mRNA, and CDS. The last column only includes
ID name like ’ID=XP00000’ (see below).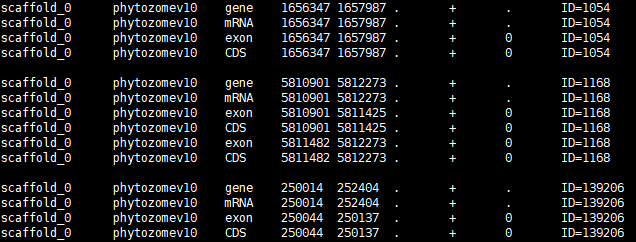
-
RESOURCE_PROTEIN in RESOURCE.config means combined target protein sequences in
multiple genomes and users should prepare them. For example, we extracted and
used protein sequences as RESOURCE PROTEIN containing FAR1 (PF3101) NBARC
(PF00931) in plant genomes as well as C2H2 zinc finger(PF00096) and
homeobox(PF00046) in animal genomes.
The IDs in protein and tsv files of target or allied species should be
less than 25 characters.
ReplyList
Register된 Reply이 없습니다.

